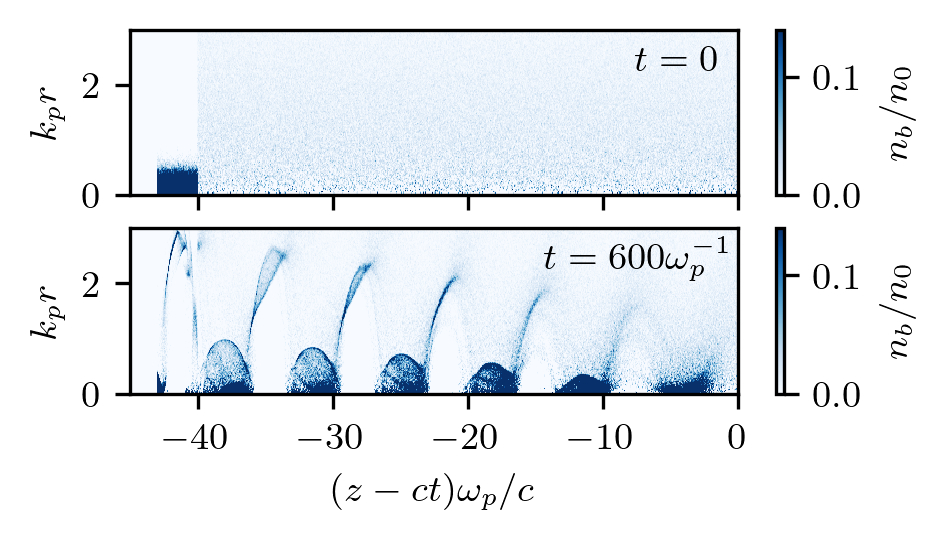
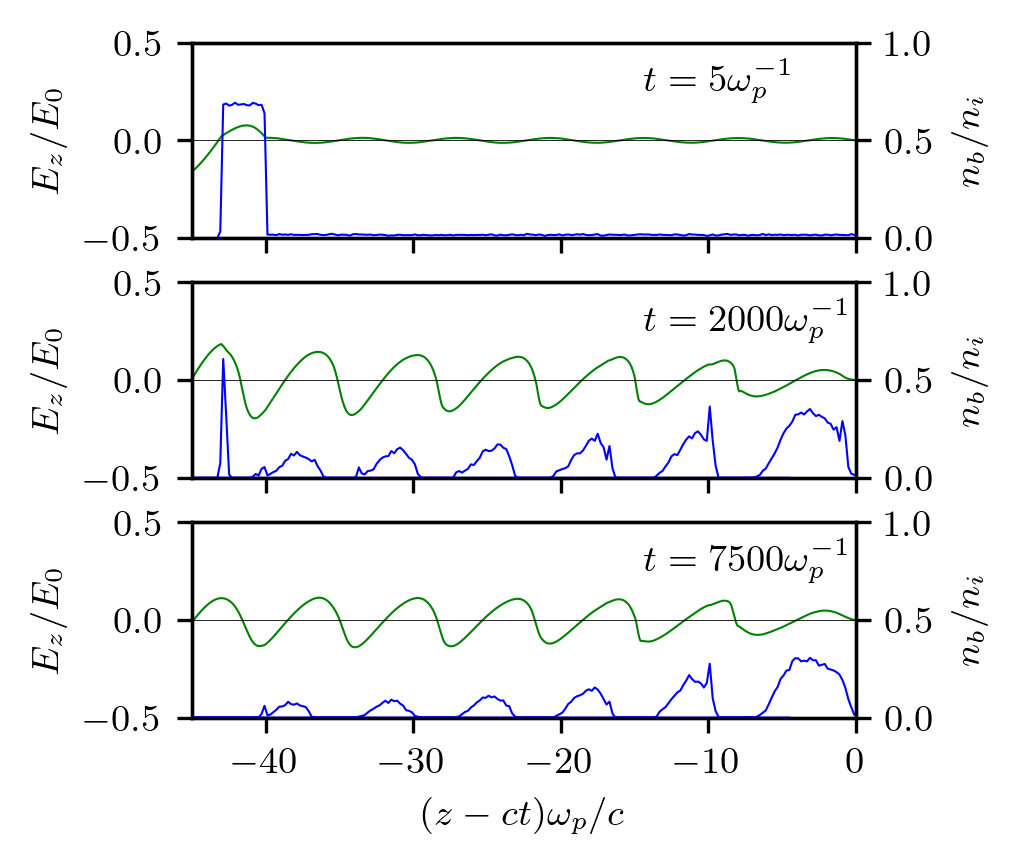
Seeded self-modulation
This is an example of seeded self-modulation of a particle beam. The run reproduces results shown in Fig.2 of the paper where the self-modulation effect was first discovered. The results do not exactly coincide with those in the paper because the original figure was produced using a fluid plasma solver and much wider beam particles.
Code launcher:
original-ssm.py.Post-processing (Jupyter notebook):
original-ssm-figs.ipynband images that it produces:
Code launcher:
from lcode.simulation import Simulation
from lcode.diagnostics import FXiDiag, FXiType, OutputType, ParticlesDiag
from lcode.diagnostics import SliceDiag, SliceType, SliceValue
# Set parameters of the solver:
config = {
'geometry': 'circ',
'processing-unit-type': 'cpu',
'window-width': 5,
'transverse-step': 0.02,
'window-length': 45,
'xi-step': 0.02,
'time-limit': 7500.1,
'time-step': 5,
'plasma-particles-per-cell': 10,
'ion-model': 'background',
}
# Set beams
beams = {
'current': -0.03, 'particles_in_layer': 250,
'default': {'angspread':2e-4, 'energy': 1000},
'driver': {'xishape':'l', 'length': 40, 'radius': 2},
'witness': {'xishape':'l', 'length': 3, 'radius': 0.2}
}
# Set diagnostics
diag = [FXiDiag(
output_period=100,
output_type=OutputType.NUMBERS,
f_xi=FXiType.Ez | FXiType.rho_beam),
SliceDiag(
SliceType.XI_X,
output_period=100,
output_type=OutputType.NUMBERS,
slice_value = SliceValue.rho_beam | SliceValue.ne)
]
sim = Simulation(config=config, diagnostics=diag,
beam_parameters=beams)
sim.step()